My thoughts about supposed haplotype of Ramesses III and its prediction.
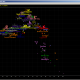
At least twice we received emails asking about NevGen’s prediction of haplotype of Ramesses III. His haplotype is supposed to be this, in NevGen format: 8,21,19,8,20-20,0,0,0,13,17,33,0,0-0,0,0,0,14,20,0,0-0-0-0,0,13,0-0,13,0,0,0,0-0,0,10 Predicting it in General Level of predictor I got this: Picture I got for top haplogroup (J1a2a1a2 P58) looks like this: You can see most of values look like being out of normal, by...