2019-2020 testing of prediction, from August 31st 2019 to June 14th 2020
Here are new results of testing of NevGen Predictor, of last 10 and half months.
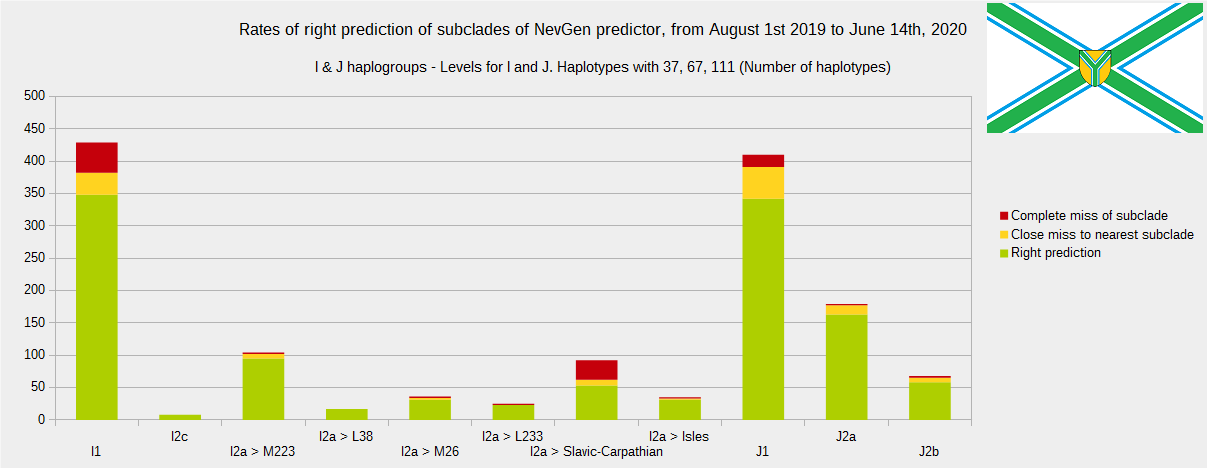
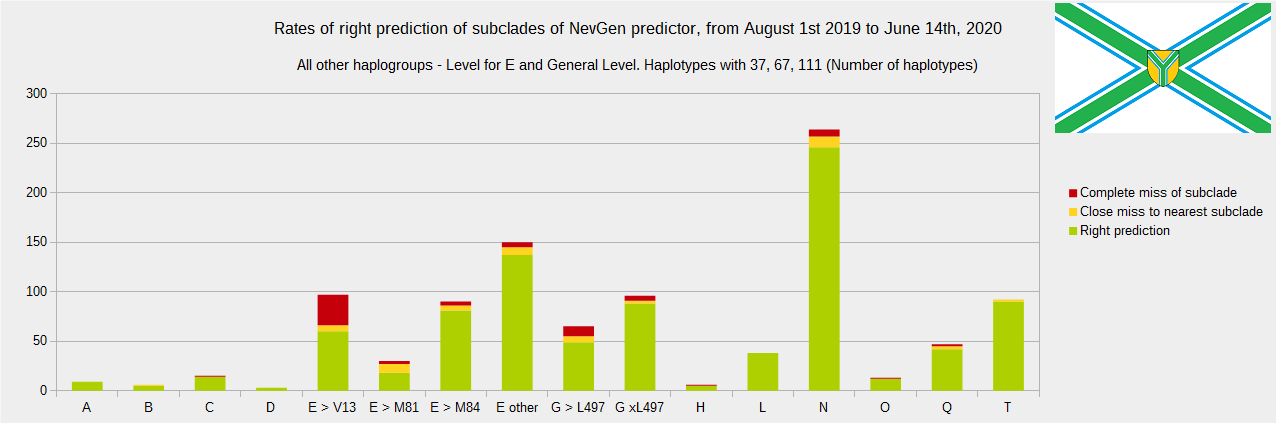
Newly found haplotypes with known deep SNPs are predicted for (their already known) subclade with NevGen Predictor. Statistics of results you can see on the next six pictures (three are by number of haplotypes used, and other three by percentages). We need to say that haplotypes which do not belong to any of supported subclades (due to small number of available samples) are not used in this statistics.
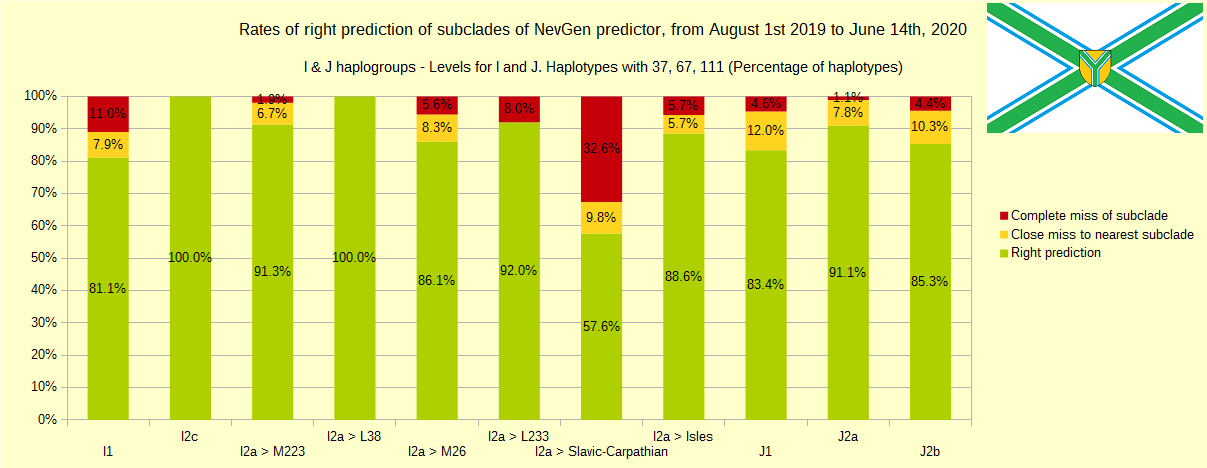
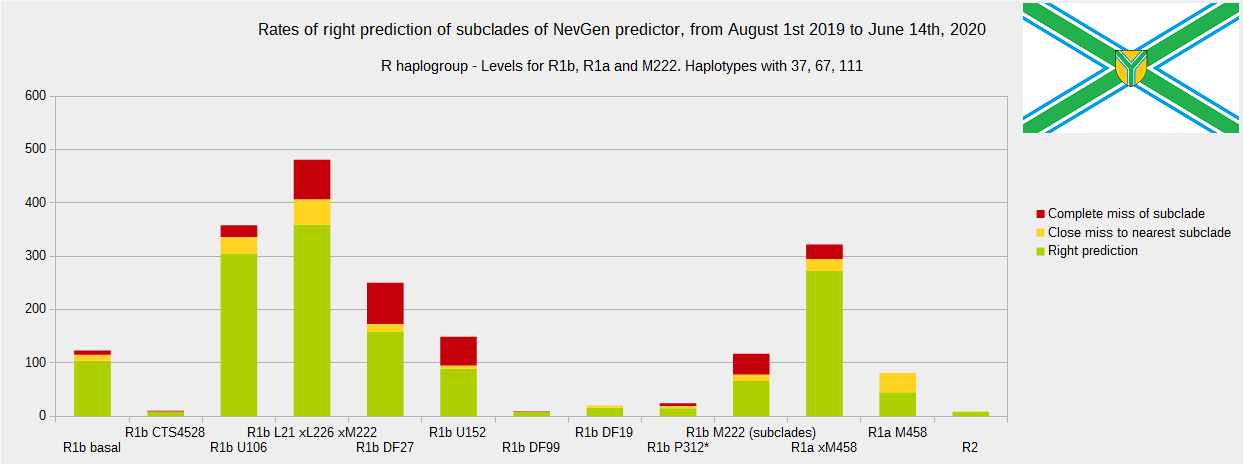
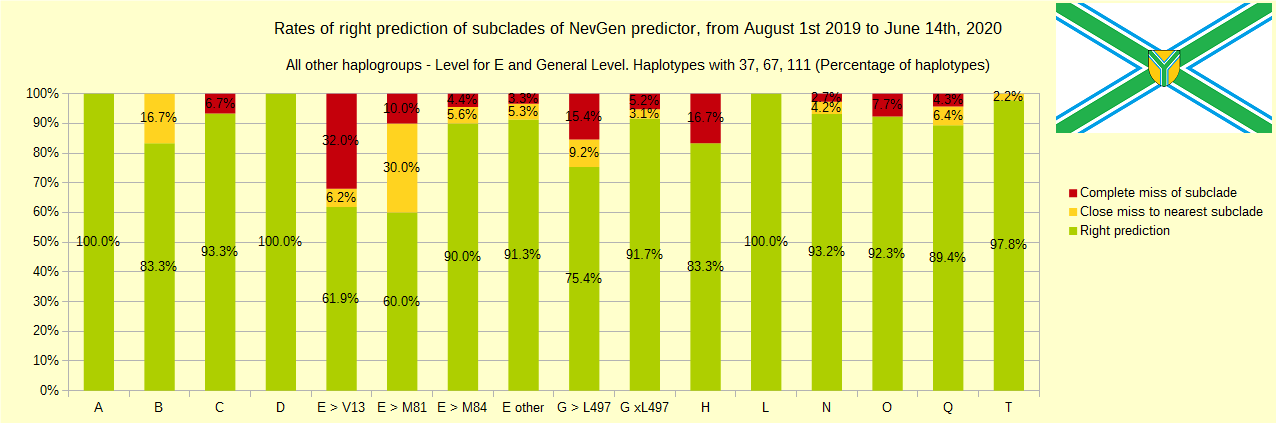
First is for haplogroups R1b, R1a and R2, second is for haplogroups I and J, and third is for all other haplogroups. As we can see, percentages of right predictions are similar to prediction rates of July 31st 2019.
Like we said before, to avoid confusion, should be noted that statistics is concerned with prediction of deeper subclades of mentioned higher-level haplogroups, not for prediction of higher-level haplogroups themselves. For example, I1’s 81% of right subclade predictions is for prediction of subclades of I1, not for prediction of I1 alone, which is easy with at least 25 markers. The same holds for all other haplogroups from pictures, like I2a > L38, I2a > Slavic-Carpathian, I2a > Isles, I2a > M223, J1, E > V13, R1b > M222, R1a M458, C, L, Q, T, D and so on.
Important thing is that all haplotypes which are less than 9/111, 6/67 or 4/37 close to any haplotype which is part of statistics of it’s already known subclade are excluded from this statistics. That way statistics is more reliable, since such close haplotypes are almost always good predicted. Haplotypes with less than 37 markers were not used in this testing, but majority of them had 111 markers.
Level for R1b-M222 used here is not available in public, since we are not satisfied with it’s results.
From statistics for R1b-L21 are excluded all haplotypes of M222 and L226, because they are trivial to predict themselves (but not their deeper subclades).
As we can see, this year again the worst prediction results are for R1b-U152, 59.7% (but somewhat better than last year’s 47.3%, due to better haplotype training set). I still believe we need many hundreds (if not thousands) of new haplotypes to get satisfying results. Right prediction rate of R1b-U106 subclades is slightly down from 85.1% to 84.9%, and wrong prediction rate is up from 5.4% to 6.1%, probably due to addition of new U106 subclades into R1b Level with low number of haplotypes in training dataset.
Like in the last year, with yellow colour we have marked haplotypes which got the most of probability to subclade which is very close, one level below. For example, if our haplotype is by SNP proven to belong to “R1b U106>Z381> Z301>L48> Z9>>Z326>> A5011”, but we got top probability in NevGen for “R1b U106>Z381> Z301>L48> Z9>>Z326>> BY4305”, we than record this testing as “close miss”.
From our experience, subclades under L1335 and FGC11134 (R1b > L21), are not easy to be distinguished, and also Z326 under U106 or L1029 (R1a > M458) or under L258 (I1), just to mention some of them.
Now something we never wrote about before: long-distance right predictions. That are predictions when haplotypes were right predicted, despite being very far from any sample in our statistics of it’s subclade. I started to record such cases about 18 months ago, and till now I have 98 of them, when distance from nearest is at least 30/111. Such cases mostly happen in haplogroups E and J2a.
Our most distant successful prediction is for 111-markers haplotype of haplogroup G2b M3155, which is right predicted despite being 78 markers far from its nearest in training dataset for its haplogroup (it was predicted with 33.29% probability).
Second most distant successful prediction is in haplogroup J2a > Z6048, where distance to nearest was 71. In R1b, the greatest distance with right prediction was 41, and haplotype belonged to subclade R1b DF27>Z196> L176.2>CTS4188> S11121.
Here you can see whole list of such long-distance right predictions, by haplogroup, with distance from nearest, and in many cases it’s subclade.
E (24 haplotypes)
Distances: 37/111 (subclade of V13), 49 (E1b1b V22), 42, 41, 48, 46 (M84), 43, 47 (M84), 42, 42, 40 (M84), 41 (E1b1a V38>> L485), 40 (E1b1b PF1975), 43 (E1b1b V22>> PH2818> BY1984), 46 (E1b1b M123> PF4428), 40 (E1b1b V1515), 40 (E1b1b V1515> V1700), 55, 53, 52, 52 (E1b1a V38>> M4231), 50 (E1b1b M123>M34> M84>> PF6751> PF6748), 61 (E1a M132), 67 (E1b1b V1515)
J2a (19 haplotypes)
39, 37, 39, 32, 41, 46, 40, 41, 41, 42, 42, 41, 44, 46 (J2a1 Z7700> FGC9883), 50 (J2a1 Z7700> FGC9883), 52 (J2a1 Z387), 54 (J2a1 PF5191>> S15439), 63 (J2a1 M319), 71 (Z6048)
R1b (12 haplotypes)
35 (R1b DF27>Z196> Z209> Z295> Z216>> PH1171), 33 (R1b Z2103> Z2106), 33 (R1b > PF7589), 31(R1b U106>Z381> Z301>>Z30>> S22165), 34 (R1b L21>DF13> DF21>FGC3213> S3058),
32 (R1b U106>Z381> Z156>DF98> S1911), 33 (R1b DF27>ZZ12> ZZ19>Z31644> A2146), 30 (R1b DF27>Z196> Z272>DF17), 36 (R1b U106>Z381> Z156> FGC39801), 40 (R1b L21>DF63> BY592), 40 (R1b V88 >> V1589),
41 (R1b DF27>Z196> L176.2>CTS4188> S11121)
C (6 haplotypes)
45, 57 (C2 M217> F1067), 54, 54, 54 (C2 M217> F1067), 63 (C2 M217> F1067)
L (6 haplotypes)
42, 45, 47, 51 (L1b M317), 59 (L1b M317> M349), 60 (L1b M317> M349)
Q (5 haplotypes)
47 (Q M346>> L330), 45, 49 (Q M346>> Y4800> F835> L932), 46 (Q M346>> Y4800> F835> L932), 50 (Q M346>> Y4800> F835> L932)
G (5 haplotypes)
47 (G2a2a PF3147> Z36520), 49 (G1a CTS11562), 44 (G2a2 >> Z30503), 52 (G1a CTS11562), 78 (G2b M3155)
T (5 haplotypes)
39, 49 (T > Y11151), 41, 41, 43 (T L131)
O (4 haplotypes)
47 (O1b1 F2320), 46 (O2a2 F525), 44 (O2a1 F51), 62 (O2a2 F525)
M223 (3 haplotypes)
32 (I2a2a M223>Y4450>> M284>>Y3709), 32 (I2a2a M223>Z161>L801>CTS6433> L1425), 42 (I2a2a M223>S9403)
J1 (2 haplotypes)
31 (J1a >> P58>> BY111), 31 (J1a >> P58>> ZS3668)
I2a M26 (2 haplotypes)
43 (I2a1a Sardinian M26>PF4088), 41 (I2a1a Sardinian M26>PF4088)
J2b (1 haplotype)
55 (J2b >> Z2444)
A (1 haplotype)
63 (A1b1b2b M13)
I1 (1 haplotype)
40 (I1 >> Z131)
R1a (1 haplotype)
32 (YP556)








How can I contact you? There are a number of suggestions for deepening the prediction. For example DYS472=9
and DYS445=6 = https://www.yfull.com/live/tree/J-FGC34168/
can only get:
‘An error occurred while calculating the haplogroup. Please check the entered data.’
Tried several samples, 2 haplogroups, copy/paste STRs from FTDNA.
tried in 2 browsers and restarted computer.
I’ve used the site hundreds of times before.
Stephen Arbon
Can we add more SNP’s for R1a-Z93 if possible?