My thoughts about supposed haplotype of Ramesses III and its prediction.
At least twice we received emails asking about NevGen’s prediction of haplotype of Ramesses III.
His haplotype is supposed to be this, in NevGen format:
8,21,19,8,20-20,0,0,0,13,17,33,0,0-0,0,0,0,14,20,0,0-0-0-0,0,13,0-0,13,0,0,0,0-0,0,10
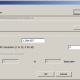
Predicting it in General Level of predictor I got this:
Probability of unsupported subclade: 100.00%
Warning: Values of fitness are too small. Probably is error in order of STRs!
Probability = 0.00% Fitness=0.43 [0.01] J1a2a1a2 P58
Probability = 0.00% Fitness=0.88 [0.02] E1b1b V22
Probability = 0.00% Fitness=0.68 [0.02] Q M346>> Z780
Probability = 0.00% Fitness=0.66 [0.01] D1a1a2 F1070
Probability = 0.00% Fitness=0.66 [0.01] A0a V148
Probability = 0.00% Fitness=0.61 [0.02] Q M346>> M3> M902
Probability = 0.00% Fitness=0.55 [0.01] I2a1 S21825>> L880 ("Northern France")
Probability = 0.00% Fitness=0.51 [0.01] Q F1096> M25
Probability = 0.00% Fitness=0.45 [0.01] O1a1 > CTS5726
Probability = 0.00% Fitness=0.44 [0.01] E1b1b > V12
Probability = 0.00% Fitness=0.43 [0.01] Q M346>> M3
.
.
.
Probability = 0.00% Fitness=0.05 [0.00] G2a2 >> M278
Probability = 0.00% Fitness=0.05 [0.00] Q M346> YP4004
Probability = 0.00% Fitness=0.04 [0.00] R1a YP1272
Probability = 0.00% Fitness=0.03 [0.00] T >> CTS11451> Y4119>> Y29991
Probability = 0.00% Fitness=0.02 [0.00] C2b1a1b1 F3985
Probability = 0.00% Fitness=0.02 [0.00] A00
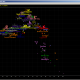
Picture I got for top haplogroup (J1a2a1a2 P58) looks like this:
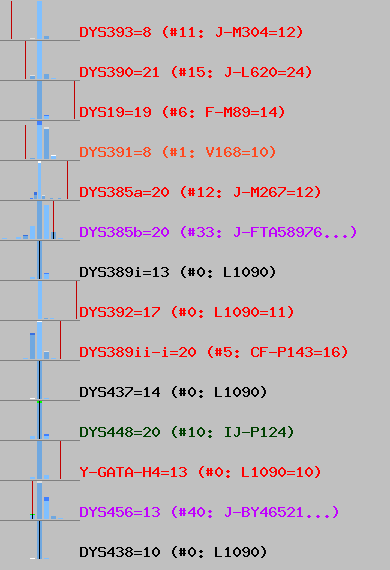
You can see most of values look like being out of normal, by position of red vertical line. It fits into it like elephant fits into glass shop.
From it, I suspected some values from this haplotype might be random (i.e. wrong). So, few days ago I wrote code which generates random haplotypes, using the same STRs which are part of Ramesses’s haplotype.
I generated two hundred such haplotypes, and average top fitness of predictions of all those random haplotypes was 0.12%, which is something lower than 0.43% for supposed haplotype of Ramesses III. Here are some samples of them:
9,21,15,9,10-21,0,0,0,16,10,23,0,0,0,0,0,11,25,0,0,0,7,0,9,0,0,0,0,0,11
15,20,13,6,16-21,0,0,0,17,19,36,0,0,0,0,0,15,23,0,0,0,22,0,22,0,0,0,0,0,12
11,28,12,12,7-11,0,0,0,14,17,35,0,0,0,0,0,20,16,0,0,0,22,0,17,0,0,0,0,0,14
14,20,13,10,10-20,0,0,0,13,15,25,0,0,0,0,0,11,20,0,0,0,14,0,22,0,0,0,0,0,7
17,21,18,7,12-20,0,0,0,15,16,24,0,0,0,0,0,21,17,0,0,0,14,0,22,0,0,0,0,0,12
13,28,13,14,10-20,0,0,0,17,13,21,0,0,0,0,0,17,17,0,0,0,24,0,18,0,0,0,0,0,15
10,22,11,7,17-22,0,0,0,11,6,44,0,0,0,0,0,12,17,0,0,0,7,0,19,0,0,0,0,0,11
13,26,13,11,16-18,0,0,0,15,8,26,0,0,0,0,0,15,9,0,0,0,7,0,22,0,0,0,0,0,15
Then I made several other sets of random haplotypes, which contain from 1 to 13 real values (randomly chosen which will be taken) from real haplotype, which belongs to E1b1a V38>> M4231, and the rest of values are random numbers.
Real haplotype, which is used to borrow values from it, is:
13,21,15,11,16-17,0,0,0,13,11,31,0,0,0,0,0,14,21,0,0,0,11,0,15,0,0,0,0,0,10
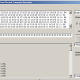
Here you can see table of average fitnesses for top prediction for all haplotypes of partially random generated haplotypes, 200 of haplotypes in each group. Grops are sorted by number of values which are taken from real haplotype.
0 0.12% (200 random haplotypes - All 14 values are random)
1 0.18% (200 haplotypes - 1 value is from real haplotype, 13 other values are random)
2 0.33% (200 haplotypes - 2 values are from real haplotype, 12 other values are random)
3 0.58% (200 haplotypes - 3 values are from real haplotype, 11 other values are random)
4 0.90% (200 haplotypes - 4 values are from real haplotype, 10 other values are random)
5 1.57% (200 haplotypes - 5 values are from real haplotype, 9 other values are random)
6 2.52% (200 haplotypes - 6 values are from real haplotype, 8 other values are random)
7 3.56% (200 haplotypes - 7 values are from real haplotype, 7 other values are random)
8 5.04% (200 haplotypes - 8 values are from real haplotype, 6 other values are random)
9 7.12% (200 haplotypes - 9 values are from real haplotype, 5 other values are random)
10 9.94% (200 haplotypes - 10 values are from real haplotype, 4 other values are random)
11 13.78% (200 haplotypes - 11 values are from real haplotype, 3 other values are random)
12 20.26% (200 haplotypes - 12 values are from real haplotype, 2 other values are random)
13 31.03% (200 haplotypes - 13 values are from real haplotype, 1 other value is random)
14 52.13% (All 14 values are from real haplotype)
From table you can see that top fitness of supposed Ramesses’s haplotype (0.43%) fits between average fitnesses (0.33% and 0.58%) with 2 and 3 real values and all other random values.
So, it seems to me that most probably only 2-4 values of Ramesses’s haplotype are real, and all others are wrong (random) values. Unless Ramesses III belonged to major haplogroup which is not present in modern humanity. Or is very, very rare. If Ramesses III has been not proven by SNPs to belong to E1b1a, from his STRs I can not deduce anything for certain.
On youtube is video about NevGen Predictor’s prediction of Ramesses haplotype.
https://www.youtube.com/watch?v=zlrQ33MtO7Q – Reevaluating Ramesses III & Unknown Man E Y-STR Markers Using NevGen Haplogroup Predictor
I must renounce prediction on Level R1b, which assigns nonzero percentage to some of R1b subclades. It is because of imperfection of R1b Level to work with short haplotypes, due to technical reasons. It simply misses statistics (server cannot load it) for small number of markers, which enables probability of unsupported subclades to be calculated. Because of it, for this “bikini” haplotype 100% of probability is distributed to some subclades of R1b, which is not meaningful result, because of too small fitnesses. That is why R1b Level is neither meant nor recommended for use with short haplotypes.